Using boto3 to access Fresh Data Pool (EODATA) on Destination Earth
In this article, you will learn how to access Fresh Data Pool (EODATA) on Destination Earth Data Lake, using boto3 (AWS SDK for Python).
DEDL Fresh Data Pool includes satellite imagery and environmental data. The former is also known as Fresh Data Pool (EODATA) (Earth Observation Data) and with boto3 library, you can use Python to access and manage these large datasets stored in AWS S3 format.
What we are going to cover
Fresh Data Pool (EODATA) Storage endpoint
S3 is an object storage service with which you can retrieve data over HTTPS using REST API. The default S3 endpoint address to work with Fresh Data Pool (EODATA) on Destination Earth is:
https://eodata.data.destination-earth.eu
Prerequisites
No. 1 Access to My DataLake Services
You need access My DataLake Services. See How to create profile on My DataLake Services.
No. 2 Credentials for S3 EODATA access from My DataLake Services
See article How to obtain eodata S3 keys through My DataLake Services
No. 3 Python
You need Python installed on your virtual machine. In this article, we are using Ubuntu 22.04 as the underlying operating system. Adjust the commands if you are using something else.
Execute this command to test whether Python is already installed or not:
python3 --version
If the reply contains version number then yes, Python is installed and ready to be used:
Python 3.12.3
To install Python on Linux, see How to install Python virtualenv or virtualenvwrapper
No. 4 boto3 library installed
If you are using Python environment like virtualenv, enter the environment in which you wish to install boto3. In it, execute the following command:
pip3 install boto3
You can also install the package globally:
sudo apt install python3-boto3
How to create executable Python files for this article
Use nano
Make sure you have an editor installed to create Python files with it. For example, install nano text editor with a command such as
sudo apt install nano
To create a new file (or to edit an existing file), execute a command like this:
sudo nano boto3_eodata_browse.py
Paste the script that you want to execute (this list is below). Perform appropriate modifications to the code as instructed (like assigning values to variables). Save the file.
Once you have exited from the text editor, execute python3 command followed by the name of your script from the directory it is in. For example:
python3 boto3_eodata_browse.py
The script in file boto3_eodata_browse.py should be executed.
Note
For simplicity sake, in this article we will be executing only Python scripts from the folder that contains its file. Be sure to use a command such as cd to move focus to that folder or adjust files addresses in the scripts below.
Scripts developed in this article
Here is a list of Python scripts in this article:
- boto3_eodata_browse.py
Browse the Fresh Data Pool (EODATA) repository
- boto3_eodata_browse_list.py
Listing the downloaded data by Prefix
- boto3_eodata_download_single_file.py
How to download a PNG file
- boto3_eodata_eof.py
Downloading .EOF file
- boto3_eodata_download_single_tiff.py
Downloading a .TIFF file from Fresh Data Pool (EODATA) repositories
- boto3_eodata_safe.py
How to download a .SAFE directory
Browsing the Fresh Data Pool (EODATA)
You can use boto3 to browse the Fresh Data Pool (EODATA) repository. This is the Python code you are going to use:
boto3_eodata_browse.py
import boto3
access_key='YOUR_ACCESS_KEY'
secret_key='YOUR_SECRET_KEY'
directory='Envisat-ASAR/ASAR/ASA_WSS_1P/2012/04/08/'
host='https://eodata.data.destination-earth.eu'
container='EODATA'
s3=boto3.client('s3',aws_access_key_id=access_key, aws_secret_access_key=secret_key,endpoint_url=host)
print(s3.list_objects(Delimiter='/',Bucket=container,Prefix=directory,MaxKeys=30000)['CommonPrefixes'])
These are the variables used in the code:
- access_key
Your access key. Obtain it by following Prerequisite No. 2.
- secret_key
Your secret key. Obtain it by following Prerequisite No. 2.
- directory
The directory within Fresh Data Pool (EODATA) repository which you want to explore.
When filling in the variable directory, make sure to follow these rules:
Use slashes / as separators between elements of that path - directories and files
Do not start the path with a slash /
Since the element you are exploring is a directory, finish the path with a slash /
Start path with folder name found within the root directory of the Fresh Data Pool (EODATA) repository (for example Sentinel-2 or Sentinel-5P)
If you want to explore the root directory of the Fresh Data Pool (EODATA) repository, assign an empty string to variable directory:
directory=''
If you don’t have a directory which you want to explore but you want to simply test this method, you can leave the value which was assigned to variable directory in the example code from above.
- host
Fresh Data Pool (EODATA) endpoint, it is always https://eodata.data.destination-earth.eu.
- container
The name of the container used.
Execute the code in file boto3_eodata_browse.py:
python3 boto3_eodata_browse.py
If you provided your access and secret keys but did not modify the variable directory, the code above will list products found in Envisat-ASAR/ASAR/ASA_WSS_1P/2012/04/08/ directory of the Fresh Data Pool (EODATA) repository. The output should look like this:
[
{
"Prefix": "Envisat-ASAR/ASAR/ASA_WSS_1P/2012/04/08/ASA_WSS_1PNESA20120408_110329_000000603113_00267_52867_0000.N1/"
},
{
"Prefix": "Envisat-ASAR/ASAR/ASA_WSS_1P/2012/04/08/ASA_WSS_1PNESA20120408_110428_000000603113_00267_52867_0000.N1/"
},
{
"Prefix": "Envisat-ASAR/ASAR/ASA_WSS_1P/2012/04/08/ASA_WSS_1PNESA20120408_110446_000000603113_00267_52867_0000.N1/"
}
]
Listing the downloaded data by Prefix
This output can be described as a “list of dictionaries”. Each of those dictionaries contains a key called Prefix, providing the path to a file or directory. Instead of printing this list like above, you can loop through it to increase the legibility of the output:
boto3_eodata_browse_list.py
import boto3
access_key='YOUR_ACCESS_KEY'
secret_key='YOUR_SECRET_KEY'
directory='Envisat-ASAR/ASAR/ASA_WSS_1P/2012/04/08/'
host='https://eodata.data.destination-earth.eu'
container='EODATA'
s3=boto3.client('s3',aws_access_key_id=access_key, aws_secret_access_key=secret_key,endpoint_url=host)
for i in s3.list_objects(Delimiter='/',Bucket=container,Prefix=directory,MaxKeys=30000)['CommonPrefixes']:
print(i['Prefix'])
Execute the code in file boto3_eodata_browse_list.py:
python3 boto3_eodata_browse_list.py
This time, the output should show only the paths:
Envisat-ASAR/ASAR/ASA_WSS_1P/2012/04/08/ASA_WSS_1PNESA20120408_110329_000000603113_00267_52867_0000.N1/
Envisat-ASAR/ASAR/ASA_WSS_1P/2012/04/08/ASA_WSS_1PNESA20120408_110428_000000603113_00267_52867_0000.N1/
Envisat-ASAR/ASAR/ASA_WSS_1P/2012/04/08/ASA_WSS_1PNESA20120408_110446_000000603113_00267_52867_0000.N1/
PNG files from a Fresh Data Pool (EODATA) repository
Basic info on PNG files
png or PNG stands for Portable Network Graphics. This image format is widely used for raster graphics; it can
store pixel data,
include transparency support and
is known for its lossless compression.
In Fresh Data Pool (EODATA), files of type .png are sometimes used for displaying visual representations of processed EODATA, such as false-color images or classification results. They do not typically include geospatial metadata like .tiff or .SAFE but may be used to show EO imagery in a more accessible, compressed format.
How to download a PNG file
The script below should download a PNG file to a directory from which the script is being executed. If that directory already contains a file which has the same name as the one you are downloading, it will be overwritten without prompt for confirmation.
The code is in file:
boto3_eodata_download_single_file.py
import boto3
access_key='YOUR_ACCESS_KEY'
secret_key='YOUR_SECRET_KEY'
key='Landsat-5/TM/L1T/2011/11/16/LS05_RMPS_TM__GTC_1P_20111116T100042_20111116T100111_147386_0194_0035_4BF1/LS05_RMPS_TM__GTC_1P_20111116T100042_20111116T100111_147386_0194_0035_4BF1.BP.PNG'
host='https://eodata.data.destination-earth.eu'
container='EODATA'
s3=boto3.resource('s3',aws_access_key_id=access_key,
aws_secret_access_key=secret_key, endpoint_url=host,)
bucket=s3.Bucket(container)
filename=key.split("/")[-1]
bucket.download_file(key, filename)
The variables are used in this piece of code is practically the same as previously. Execute with command
python3 boto3_eodata_download_single_file.py
If provided your access key and secret key but you did not change the contents of variable key, the code should download the file called
LS05_RMPS_TM__GTC_1P_20111116T100042_20111116T100111_147386_0194_0035_4BF1.BP.PNG
which is located within the root directory of product
LS05_RMPS_TM__GTC_1P_20111116T100042_20111116T100111_147386_0194_0035_4BF1
After executing the script, the output should be empty. Regardless, the downloaded file should be visible within the directory from which the script was executed. For example, this is what it will look like on Linux:

And here is what the downloaded image looks like:
EOF files from a Fresh Data Pool (EODATA) repository
What is a .EOF file
eof is short for Earth Observation Format. It is a custom format used to store and share EODATA, though it’s less common than formats like .tiff or .SAFE. Commonly used in environmental and Earth sciences, both in research and commercial EODATA systems alike. May require specialized software for reading and processing.
Internally, it is a binary or structured file format that typically includes both image data and metadata about the observation; it usually stores both raw and processed data for specific types of EO sensors or projects.
Downloading .EOF file
Put the following code into file
boto3_eodata_eof.py:
import boto3
import os
session = boto3.session.Session()
s3 = boto3.resource(
's3',
endpoint_url='https://eodata.data.destination-earth.eu',
aws_access_key_id="your_aws_access_key_id",
aws_secret_access_key="your_aws_secret_access_key",
region_name='default'
) # generated secrets
def download(bucket, product: str, target: str = "") -> None:
files = bucket.objects.filter(Prefix=product)
if not list(files):
raise FileNotFoundError(f"Could not find any files for {product}")
for file in files:
os.makedirs(os.path.dirname(f"{target}{file.key}"), exist_ok=True)
if not os.path.isdir(f"{target}{file.key}"):
bucket.download_file(file.key, f"{target}{file.key}")
# path to the product to download
download(s3.Bucket("eodata"), "Sentinel-1/AUX/AUX_POEORB/2023/10/03/S1A_OPER_AUX_POEORB_OPOD_20231024T080719_V20231003T225942_20231005T005942.EOF", "/your/path/to/python_eodata")
Be sure to replace your/path/to/ with your own directory path before running the code.
This code
downloads every file in bucket with provided product as prefix
raises FileNotFoundError if the product was not found
The arguments used are:
- bucket:
boto3 Resource bucket object
- product
Path to product
- target
Local catalog for downloaded files. Should end with an “/”. Default current directory.
Execute with command
python3 boto3_eodata_eof.py
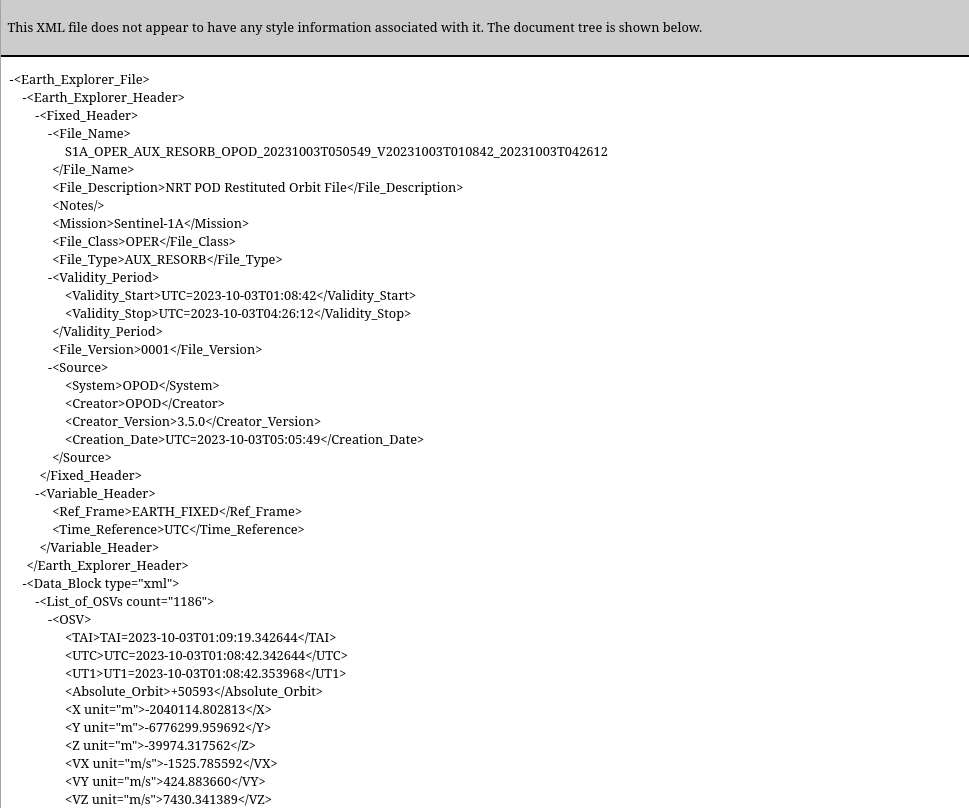
This particular file that is downloaded, is 507 Kbytes long and is containing some 15454 lines. Its start looks like this:
.TIFF files in Fresh Data Pool (EODATA) repositories
What is a .TIFF file
tiff or TIFF stands for Tagged Image File Format. It is commonly used in Earth Observation for storing satellite images. TIFF files can store geospatial metadata and large raster images, making them ideal for EODATA that require high precision.
A .tiff file consists of pixel data (images), often with associated geo-referencing data in the file header. Geospatial metadata can be stored in the GeoTIFF format (a variant of TIFF), which includes information such as the coordinate system and geo-referencing tags.
.TIFF format can store both single-band (grayscale) and multi-band (color) raster data; widely used in remote sensing applications, including vegetation mapping, land cover classification, and terrain analysis.
Downloading a .TIFF file from Fresh Data Pool (EODATA) repositories ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^6
Put the following code into file
boto3_eodata_download_single_tiff.py
import boto3
import datetime
import os
access_key='YOUR_ACCESS_KEY'
secret_key='YOUR_SECRET_KEY'
# Host and bucket info
host = 'https://eodata.data.destination-earth.eu'
container = 'EODATA'
# TIFF file inside the .SAFE directory
key = (
'Sentinel-1/SAR/SLC/2019/10/13/'
'S1B_IW_SLC__1SDV_20191013T155948_20191013T160015_018459_022C6B_13A2.SAFE/'
'measurement/s1b-iw1-slc-vh-20191013t155949-20191013t160014-018459-022c6b-001.tiff'
)
filename = key.split('/')[-1]
# Set up the S3 resource
s3 = boto3.resource(
's3',
aws_access_key_id=access_key,
aws_secret_access_key=secret_key,
endpoint_url=host
)
bucket = s3.Bucket(container)
# Print start time
start_time = datetime.datetime.now()
print(f"Starting download at: {start_time}")
# Download the file
bucket.download_file(key, filename)
# Print end time
end_time = datetime.datetime.now()
print(f"Download finished at: {end_time}")
# Calculate duration
duration = end_time - start_time
print(f"Download duration: {duration.total_seconds()} seconds")
# Print file size in MB
file_size = os.path.getsize(filename) / (1024 * 1024)
print(f"Downloaded file: {filename}")
print(f"File size: {file_size:.2f} MB")
Execute with command
python3 boto3_eodata_download_single_tiff.py
This is the textual output of the code:
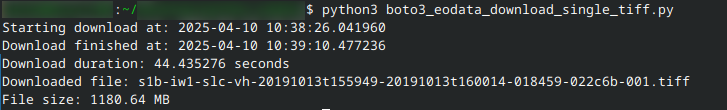
The downloaded grayscale .TIFF file looks like:

If you are downloading .SAFE directories, all .TIFF files will be automatically included into the download.
.SAFE objects in Fresh Data Pool (EODATA) repository
What is a .SAFE format
The .SAFE file format is a directory-based structure typically used by the European Space Agency (ESA) for its Sentinel satellite missions (e.g., Sentinel-1, Sentinel-2). It is not a single file but a directory structure containing metadata, image bands, calibration data, and other auxiliary files necessary for processing the satellite data. Because of that complexity, usually it requires specialized software like the ESA Sentinel Application Platform (SNAP) for proper viewing and processing.
The .SAFE product structure includes a directory containing a series of folders, each of which represents different product types, such as measurement data, calibration, metadata, etc.
Common files inside a .SAFE directory are:
- Manifest file (manifest.safe)
Contains high-level metadata about the product.
- Measurement files (e.g., .tiff, .png)
Represent the EO image data.
- Metadata files (.xml)
Contain detailed information about the acquisition, including satellite orbit data, processing level, and time stamps.
How to download a .SAFE directory
Put the following code into file
boto3_eodata_safe.py:
import boto3
import os
# Your credentials and endpoint
access_key="your_aws_access_key_id"
secret_key="your_aws_secret_access_key"
endpoint = 'https://eodata.data.destination-earth.eu'
bucket_name = 'eodata'
prefix = 'Sentinel-1/SAR/SLC/2019/10/13/S1B_IW_SLC__1SDV_20191013T155948_20191013T160015_018459_022C6B_13A2.SAFE/'
# Local directory where files will be saved
local_base_dir = './downloaded_files' # Change this to your desired folder
# Create the client
s3 = boto3.client(
's3',
aws_access_key_id=access_key,
aws_secret_access_key=secret_key,
endpoint_url=endpoint
)
# Create the local base directory if it doesn't exist
os.makedirs(local_base_dir, exist_ok=True)
# Handle pagination
continuation_token = ''
while True:
# List objects with pagination (max 1000 items per page)
list_params = {
'Bucket': bucket_name,
'Prefix': prefix,
'ContinuationToken': continuation_token if continuation_token else '' # Use empty string if no continuation token
}
response = s3.list_objects_v2(**list_params)
# Loop through each object
for obj in response.get('Contents', []):
key = obj['Key']
# Skip directories (keys that end with "/")
if key.endswith('/'):
continue
# Generate local file path, preserving folder structure
local_path = os.path.join(local_base_dir, key) # Save to specified folder
local_dir = os.path.dirname(local_path) # Extract the parent directory
# Create parent directories if necessary
os.makedirs(local_dir, exist_ok=True)
# Download the file to the local path
print(f'Downloading {key} -> {local_path}')
s3.download_file(bucket_name, key, local_path)
# Check if there are more files to list
if response.get('IsTruncated'): # More files exist, continue listing
continuation_token = response.get('NextContinuationToken')
else:
break
print("✅ Download complete.")
Execute with command
python3 boto3_eodata_safe.py
Creating directories while downloading a single file
The main problem when downloading a .SAFE directory is that S3 data look like they are placed in a directory, while they are actually not. Therefore, when downloading these files, the code has to break down the file structure and translate it to directory structure needed for actual presence on Linux or Windows operating systems. To make things more complicated, if the subdirectory is not available in advance (and most of the time it won’t be), the code needs to create the relevant subdirectories on the go. In this particular case, there is a command
local_base_dir = './downloaded_files'
which defines a start of the directory and subdirectories that will be created. (It goes without saying that you could/should change this variable in case of frequent downloading of .SAFE files.)
Files downloaded from this .SAFE project
Here is the “main” directory on local disk:
Underneath, we get the actual files downloaded:
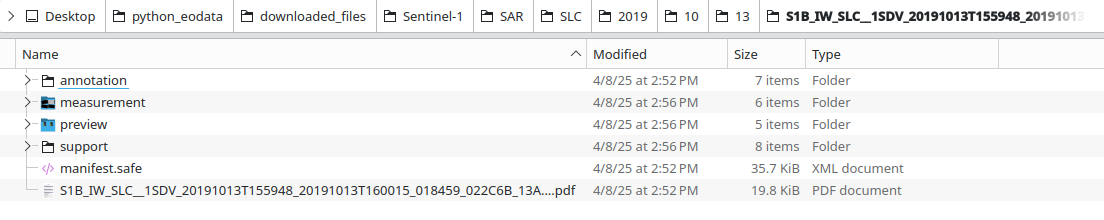
File /preview/quick-look.png shows what part of Earth surface has been observed and recorded in .SAFE format:

Under measurement subdirectory, we see the downloaded .TIFF files:
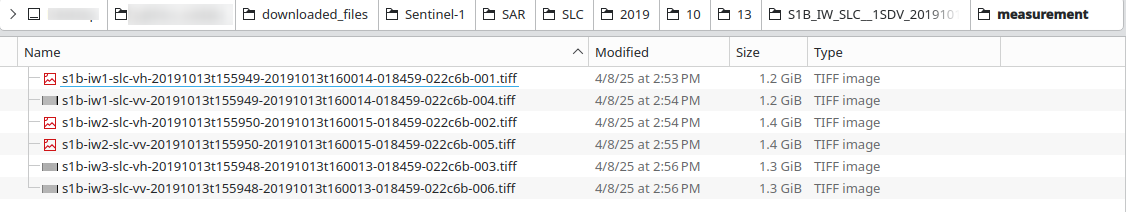
Note that .TIFF files are all longer than 1 GB of data.
There is also a PDF file for .SAFE product:

Eventual problems when downloading .SAFE files
.SAFE format is a compound of a possibly large number of possibly large files so that main problem is that files can become corrupted during the download. Also, the download may be incomplete or take too long, or the files cannot be downloaded because of their sheer size.
In conjunction, the download speeds may dwindle if you are downloading them from remote storage or if you are sharing bandwidth with others. The destination provider may also throttle your API access if you are making a large number requests while downloading files. Specifically, if you are using list_objects_vw from AWS S3 protocol, it will have a limit of 1000 objects per request.
The resolution of these problems is far beyond the context of this article, but we can also mention other possible causes of troubles:
firewall or network configuration issues,
timeouts,
invalid or missing metadata,
unrecognized or unsupported file formats,
nested directory structure handling
and so on.
What To Do Next
To access Fresh Data Pool (EODATA) on Destination Earth, you can also use s3cmd and AWS CLI:
Using AWS CLI to access Fresh Data Pool (EODATA) on Destination Earth
Using s3cmd to obtain Fresh Data Pool (EODATA) on Destination Earth